Project – Mapping Autophagy Regulation in the Gut
Autophagy is a common recycling process in which cells degrade their unnecessary or damaged parts. It plays an important role in intestinal homeostasis, and when it is not properly functioning, it can lead to gastrointestinal diseases such as Inflammatory Bowel Disease (IBD). In our lab, we are interested in studying key regulators of autophagy in different epithelial cell types in the gut in both health and disease states. This understanding may shed light on the role of autophagy in specific cell types during the development of gut diseases, and potentially lead to new targets for drug purposes. Additionally, we are interested in how autophagy in the gut is modulated by neighboring cells or commensal bacteria, and how these cell-cell interactions are disrupted during disease. To achieve our goals, we use a combination of experimental and computational approaches. In particular, we use “gut organoids” (a 3D miniature version of the human gut) from healthy or IBD patients to study autophagy regulation in different cell types and assess autophagy modulation by external factors such as microbial compounds. Additionally, we combine single-cell RNA sequencing (scRNA-seq) technologies with previously developed bioinformatics resources such as the Autophagy Regulatory Network (ARN) to the reconstruction and analyze the intracellular and intercellular signaling related to autophagy in the gut.
Autophagy is an essential stress response and degradation process in all eukaryotic cells. Autophagy plays an important role in maintaining cellular homeostasis by adapting to the changing cellular environment, cell proliferation and differentiation and the rearrangements that occur during aging. Dysregulated autophagy has been associated with various diseases such as cancer, neurodegenerative diseases and inflammatory bowel disease (IBD).
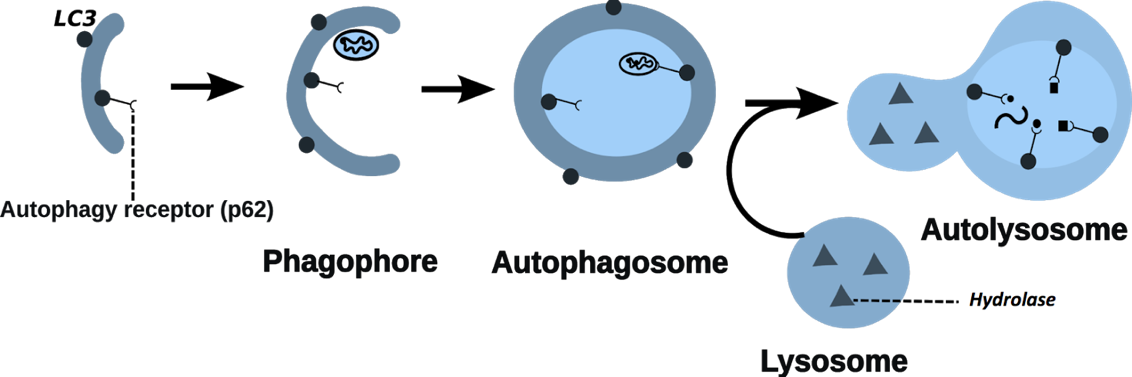
Figure 1: Process of autophagy
For full-sized version, please click on the image.
A large body of literature evidence points out the role of autophagy-related genes in the gastrointestinal tract. Autophagy is involved in the normal function of intestinal epithelial cells such as production of antimicrobial peptides by Paneth cells or mucus production by goblet cells. Disruption of these functions is known to play a role in the pathogenesis of IBD. Genome-wide association studies have revealed single nucleotide polymorphisms (SNPs) in autophagy-related genes that contribute to the susceptibility to develop IBD. However, little is known about the cell-type specificity of these mutations.
The reconstruction and analysis of protein-protein interaction networks can help to better understand the role of molecular interactions in biological function. We developed the Autophagy Regulatory Network (ARN) web resource (Turei et al, Autophagy, 2015), which provides an integrated and systems-level database for autophagy research. ARN enables users to examine autophagy more accurately concentrating on its comparison in different tissues (intestinal tissue, nerve tissue, etc.) and identification of key regulators and potential new targets in diverse autophagy types. We recently updated and upgraded this resource (Csabai et al, Autophagy, 2023) by developing AutophagyNet containing updated interaction curation and integration of over 280,000 experimentally verified interactions between core autophagy proteins and their protein, transcriptional and post-transcriptional regulators as well as their potential upstream pathway connections. AutophagyNet provides annotations for each core protein about their role: 1) in different types of autophagy (mitophagy, xenophagy, etc.); 2) in distinct stages of autophagy (initiation, expansion, termination, etc.); 3) with subcellular and tissue-specific localization. These annotations can be used to filter the dataset, providing customizable download options tailored to the user’s needs.
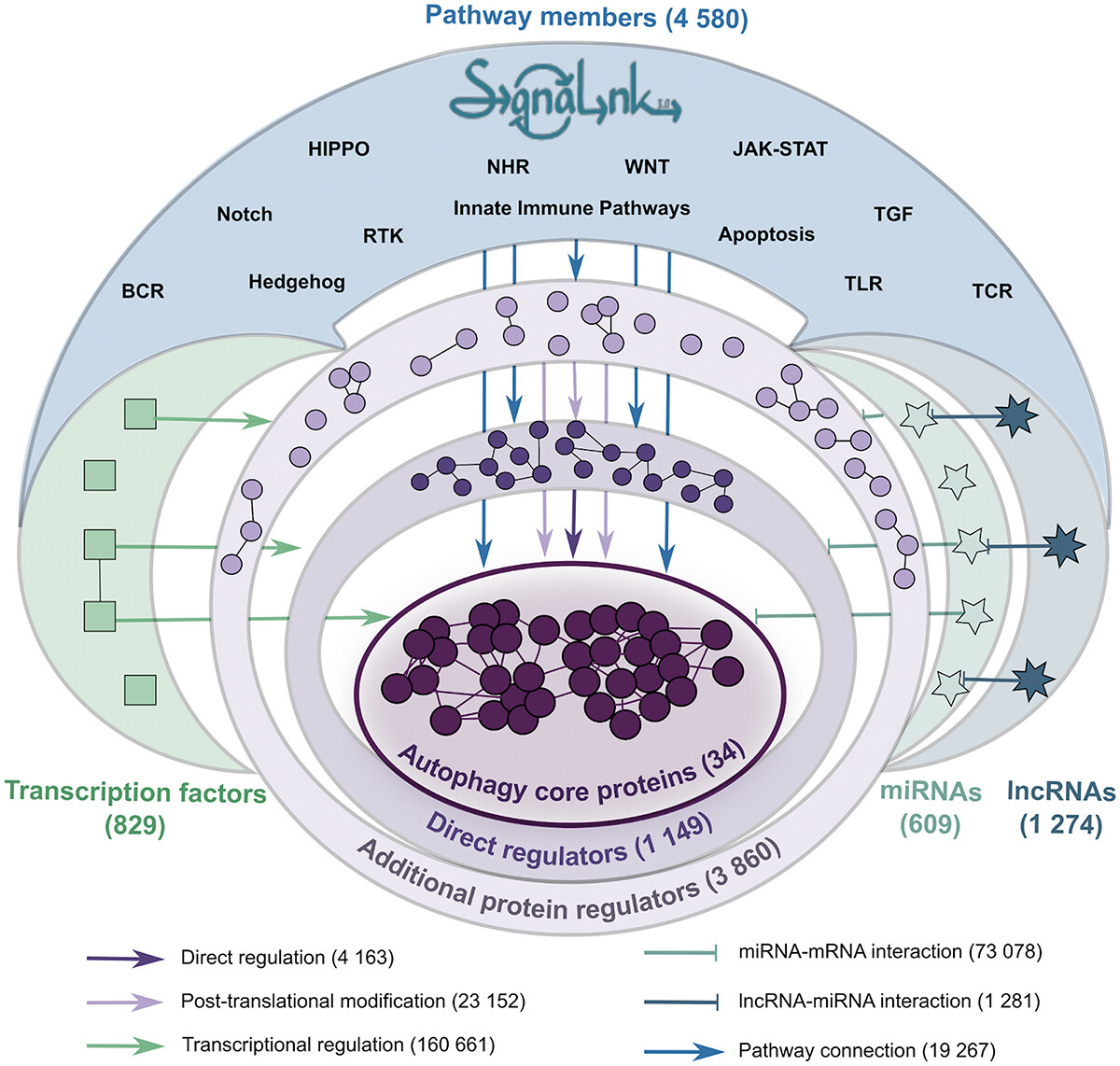
Figure 2: Autophagy regulatory interactions in Autophagy Regulatory Network 2 (AutophagyNet)
For full-sized version, please click on the image.
In recent years, with the development of single-cell RNA sequencing (scRNA-seq) technologies, an increasing number of studies have been published investigating the cell specificity of autophagy. These may shed light on the role of autophagy in specific cell types that play a major role in the development of certain diseases. While more and more is known about the cell type specificity of autophagy, the effects of cell-cell interactions on the process of autophagy remains poorly understood. It is important to understand how neighboring cells activate or reduce autophagy in each other, for example during inflammatory or infectious processes. By exploring this regulation, we can gain an even deeper understanding of disease processes and use this information to develop therapies.
Probiotic bacteria such as bifidobacteria are known to exert beneficial effects on the gut via several mechanisms of action, one of which is represented by autophagy. In our lab, we exploit human intestinal organoids from healthy or IBD donors to identify specific Bifidobacterium spp. that are able to modulate autophagy processes in the gut, and identify which epithelial cell populations are involved. In particular, we use experimental approaches (high-throughput imaging) to identify the activation of specific autophagy-related proteins activation, as well as computational approaches (network biology) to unravel signaling cascades and bacterial components through which the beneficial effects on the host are produced. With these integrated studies, we aim to identify specific strains able to restore disrupted autophagy processes in the gut. This understanding could lead to more targeted microbial therapies, which either alone or in combination with traditional approaches have the potential to revolutionize IBD management and ultimately improve patient outcomes.
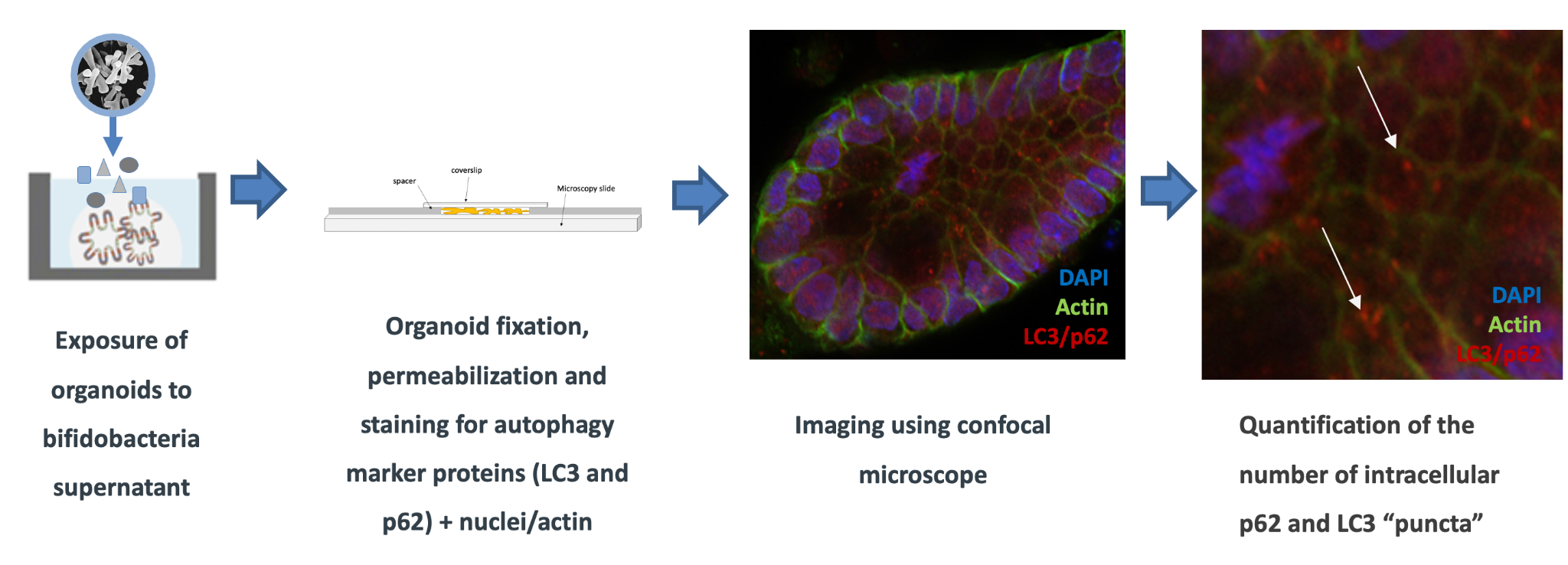
Figure 3: Imaging of autophagy proteins in organoids
For full-sized version, please click on the image.