PROJECTS
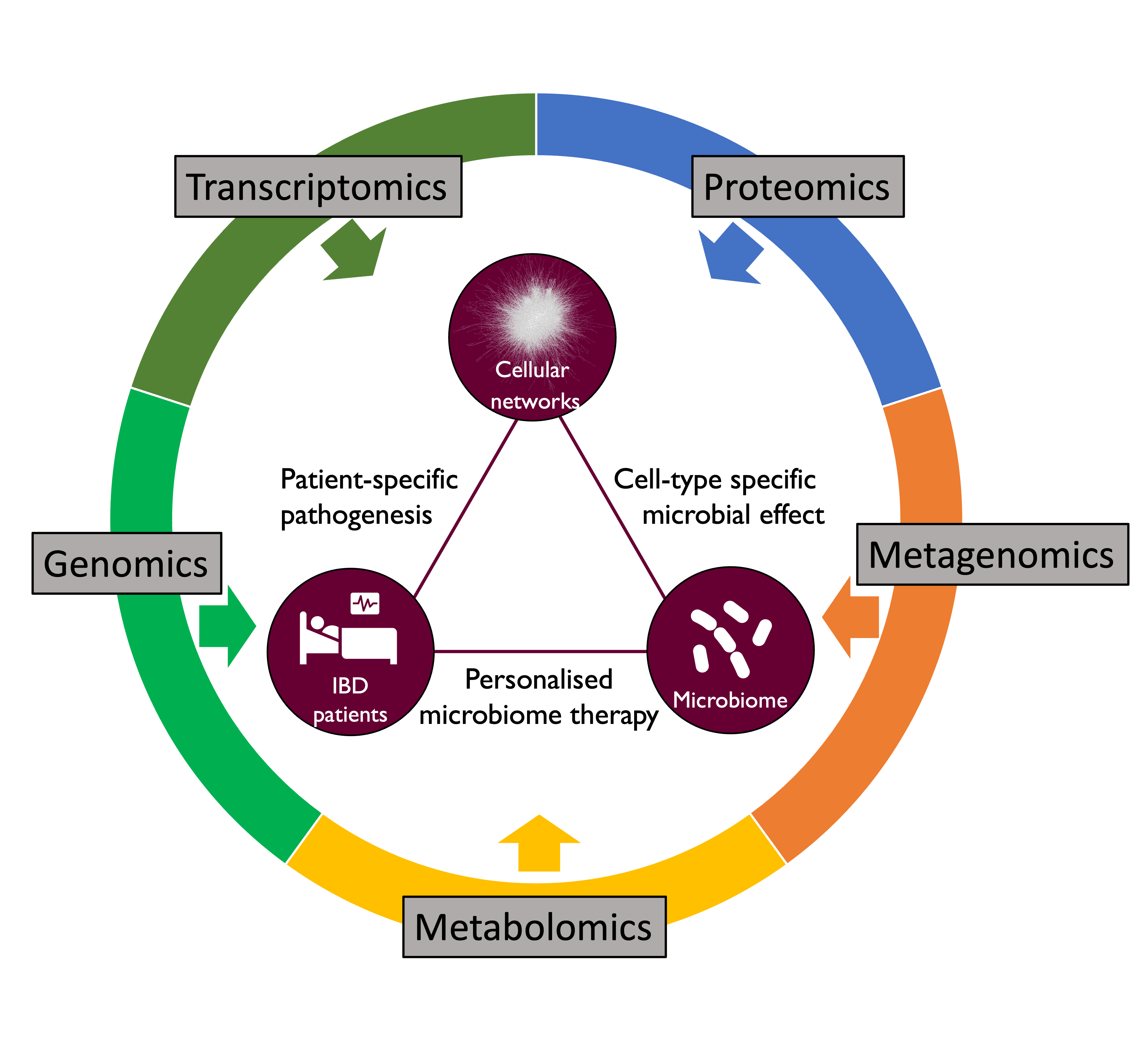
Inflammatory Bowel Disease (IBD) is a disorder of the human gastrointestinal tract characterised by inflammation of the gut mucosal layer and dysbiosis of the microbiome. IBD can broadly be classified into two diseases - Ulcerative Colitis (UC) and Crohn’s Disease (CD) - which are increasingly prevalent in the global population. However, IBD is a complex disease influenced by a variety of extrinsic and intrinsic factors such as genetics, microbiome, lifestyle (diet, smoking etc), xenobiotic exposures etc. In the Korcsmaros group, we aim to understand IBD using multiple different angles and various omics techniques as input data sources. To investigate patient-specific pathogenesis in IBD, we collaborate with Séverine Vermeire’s IBD team from Leuven and work in the UK IBD Genetic Consortia. We use patient-specific genomics and transcriptomics information from these collaborators to decipher how patient-specific genomic background contributes to IBD.
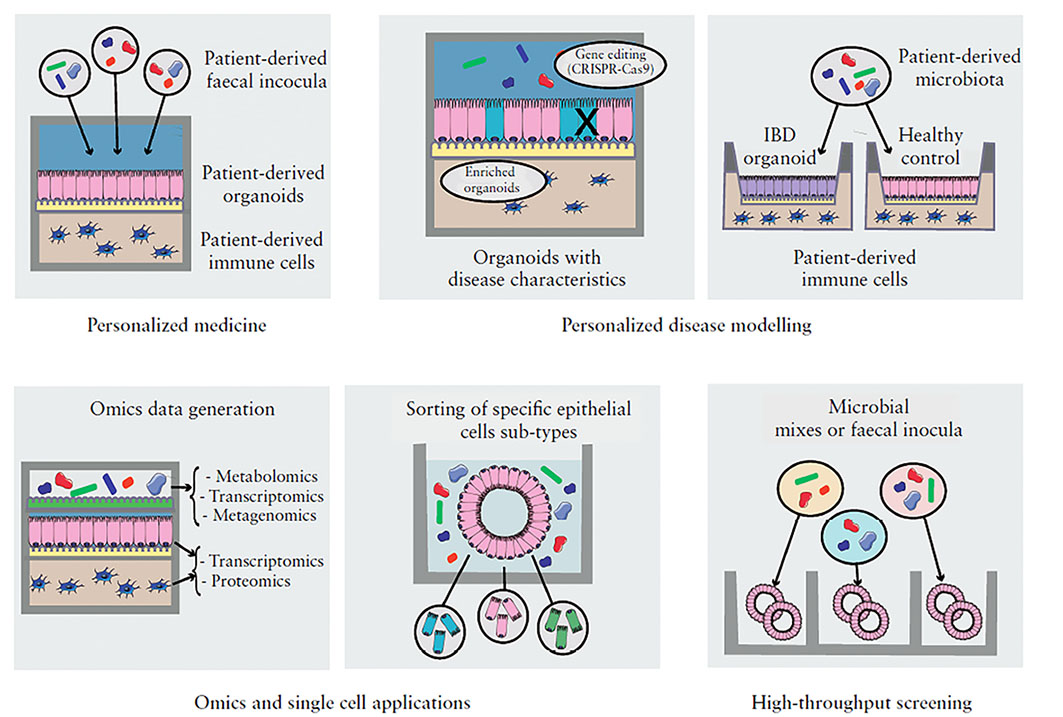
In healthy individuals, the gastrointestinal tract relies on highly sophisticated intercellular interactions to sense, adapt and respond appropriately to its cells’ immediate and distant environments. These include not only external factors (e.g. diet, resident, transitory probiotic or infectious microbes) but also other host cells from near or far distances (e.g. the underlying mesenchyme, the gut-associated innate and adaptive immune cells, the enteric nervous system, or even distant organs like the liver, the lung or the brain). Dysbiosis of the microbiota or any alteration of host cell-to-cell crosstalk can result in complex multifactorial pathologies, including Inflammatory Bowel Disease (IBD). We are combining in silico systems biology approaches with experimental data generation and validation using the human colon organoid model (colonoids). Organoids are 3D structures.
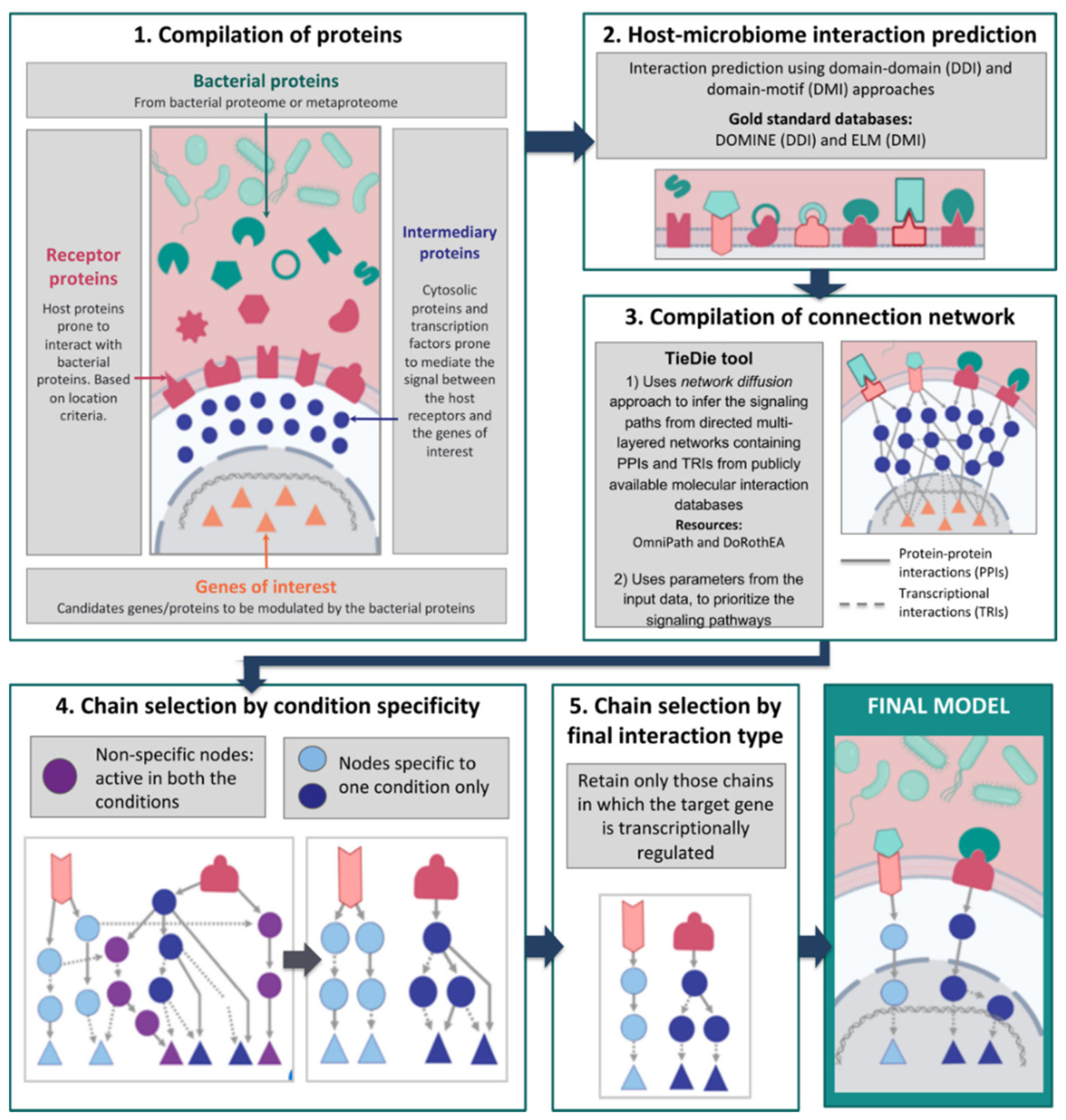
The microbiome plays a vital role in maintaining host systems' homeostatic (healthy) functions. Disrupting the healthy microbiota - by external (temperature, hygiene, etc.) or internal factors (inflammation, ageing, etc.) - causes a dysbiotic (unhealthy) state. Disruption of the microbial equilibrium between commensal and pathogen species could lead to the development of various disorders. Though differences in the microbiota between healthy and unhealthy phenotypes have been reported, the mechanistic role of the microbiota in modulating homeostatic processes remains largely unexplored. As a focused, gap-filling attempt to address this problem, we have developed in silico pipelines to predict host-microbe interactions and analyse the effect of microbes on human signalling pathways based on -omics data using network biology approaches. In industrial collaborations with BenevolentAI and Unilever, we have been testing and fine-tuning our workflows on data from the gastrointestinal tract.
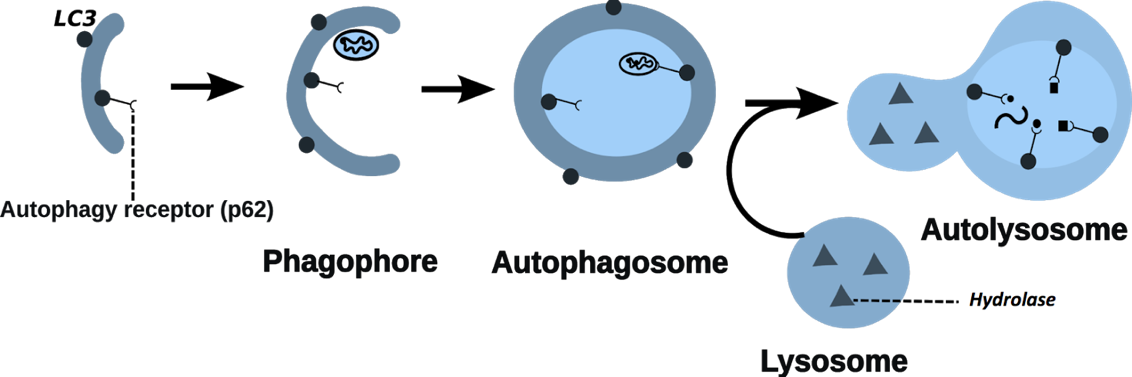
Autophagy is a common recycling process in which cells degrade their unnecessary or damaged parts. It plays an important role in intestinal homeostasis, and when it is not properly functioning, it can lead to gastrointestinal diseases such as Inflammatory Bowel Disease (IBD). In our lab, we are interested in studying key regulators of autophagy in different epithelial cell types in the gut in both health and disease states. This understanding may shed light on the role of autophagy in specific cell types during the development of gut diseases, and potentially lead to new targets for drug purposes.
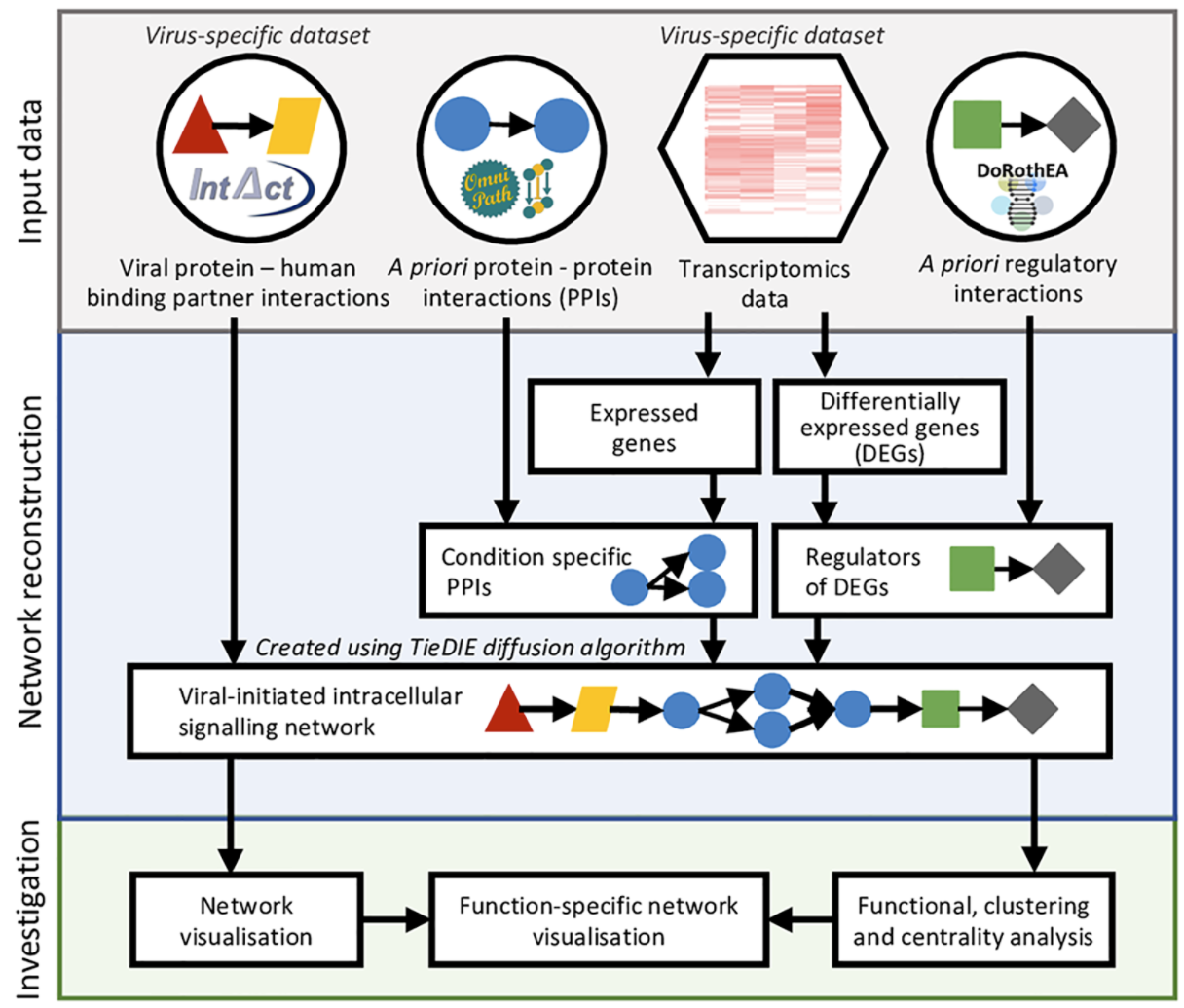
The SARS-CoV-2 pandemic has mobilised scientists around the globe to research all aspects of the coronavirus virus and the disease it causes (COVID-19). Although the pathogen primarily targets the respiratory system, its involvement in other major organs has now been documented thoroughly. To study COVID-19, we applied our expertise in multi-omics network modelling and re-utilised our resources. In particular, we have investigated how various infected cell types, especially in the gut, react to the pathogen, and how the immune response given to SARS-CoV-2 differs from other viruses. We have also developed multiple new computational tools, such as ViralLink and CytokineLink, aimed at helping the scientific community study the disease (Olbei et al, Cells, 2021, Treveil et al, PLOS Computational Biology, 2021).