Project – Multi-omics Analysis of Intestinal Organoids
In healthy individuals, the gastrointestinal tract relies on highly sophisticated intercellular interactions to sense, adapt and respond appropriately to its cells’ immediate and distant environments. These include not only external factors (e.g. diet, resident, transitory probiotic or infectious microbes) but also other host cells from near or far distances (e.g. the underlying mesenchyme, the gut-associated innate and adaptive immune cells, the enteric nervous system, or even distant organs like the liver, the lung or the brain). Dysbiosis of the microbiota or any alteration of host cell-to-cell crosstalk can result in complex multifactorial pathologies, including Inflammatory Bowel Disease (IBD). We are combining in silico systems biology approaches with experimental data generation and validation using the human colon organoid model (colonoids). Organoids are 3D structures, grown from stem cells that recapitulate the normal physiology and the different cell types of the original organ. Intestinal organoids enable the in vitro study of the regulation/dysregulation of epithelial homeostatic functions, cell-to-cell interactions and host-microbiome interactions happening in the gut during health and disease. Furthermore, the generation of multi-omics data from organoids, in combination with the application of in silico network tools, such as our OmniPath database, enables the generation of molecular interaction networks allowing contextual and multiscale analysis of the ‘omics data. These approaches facilitate deciphering molecular mechanisms involved in gut homeostasis. Complementary approaches such as high-throughput imaging platforms based at the Quadram Institute Bioscience enable us to validate experimentally the bioinformatics predictions from in silico work. By increasing the understanding of the underlying factors altered in intestinal dysbiosis-associated pathologies and the beneficial effects commensal bacteria may have on intestinal cell function, we hope to pave the way for translational developments in the prevention and treatment of gastrointestinal disorders.
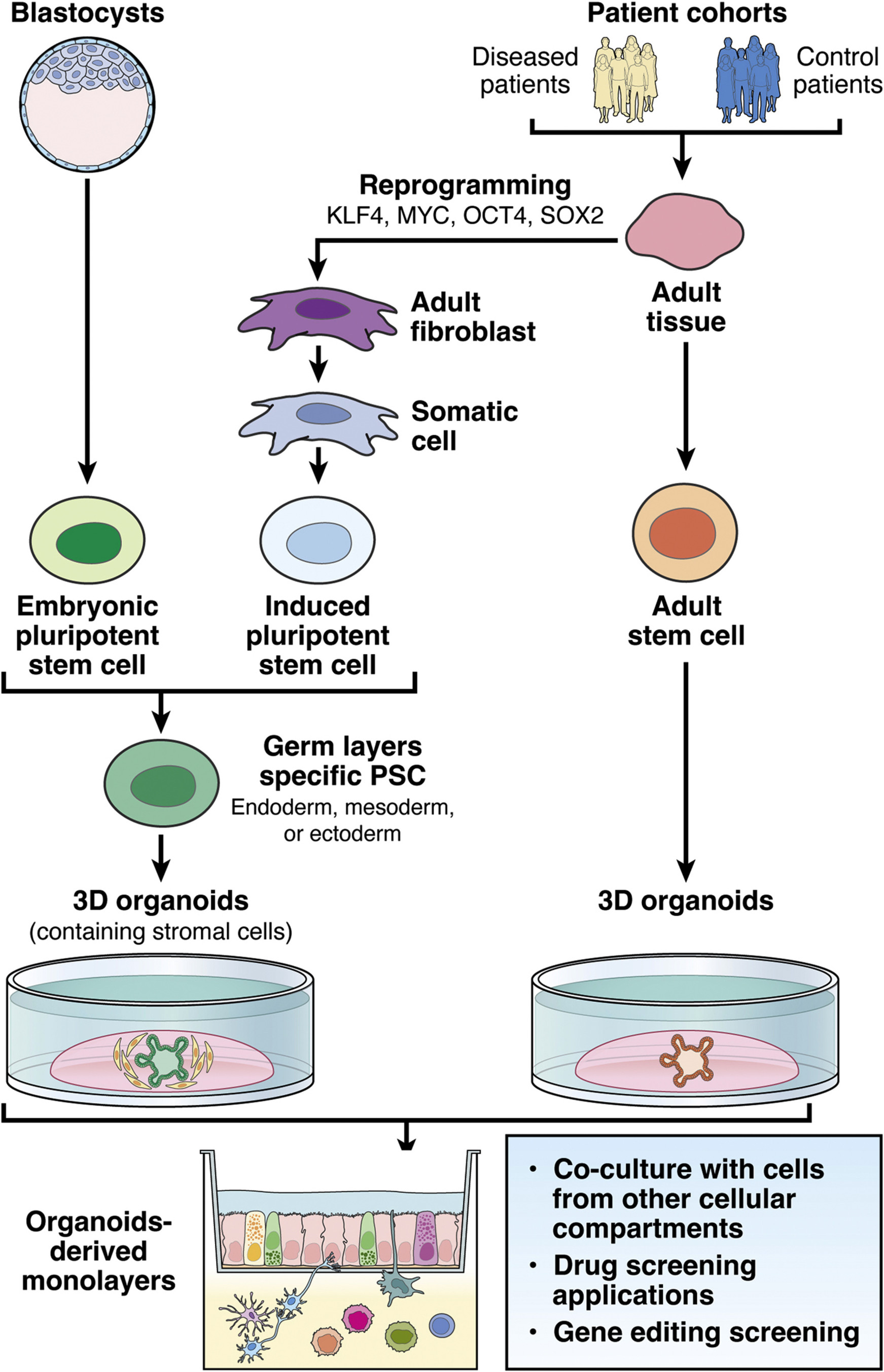
Figure 1: Our integrative research approaches to study IBD. Source: Hautefort et al, Cell Mol Gastroenterol Hepatol, 2022
For full-sized version, please click on the image.
Various signalling compounds from other organs as well as luminal factors can influence intestinal epithelial homestasis. We have chosen to separately apply commensal and probiotic Bifidobacteria strains, primary and secondary bile acids, and enteric nervous system neuromediators useto human colonoids from IBD and control patients in the culture systems illustrated below. Currently, to study the impact of various compounds listed below this paragraph the following This allows us to determine and quantify the effects each has on intestinal cell types proliferation, differentiation and function, in particular as well as the regulation of paramount cellular pathways and processes such as autophagy:
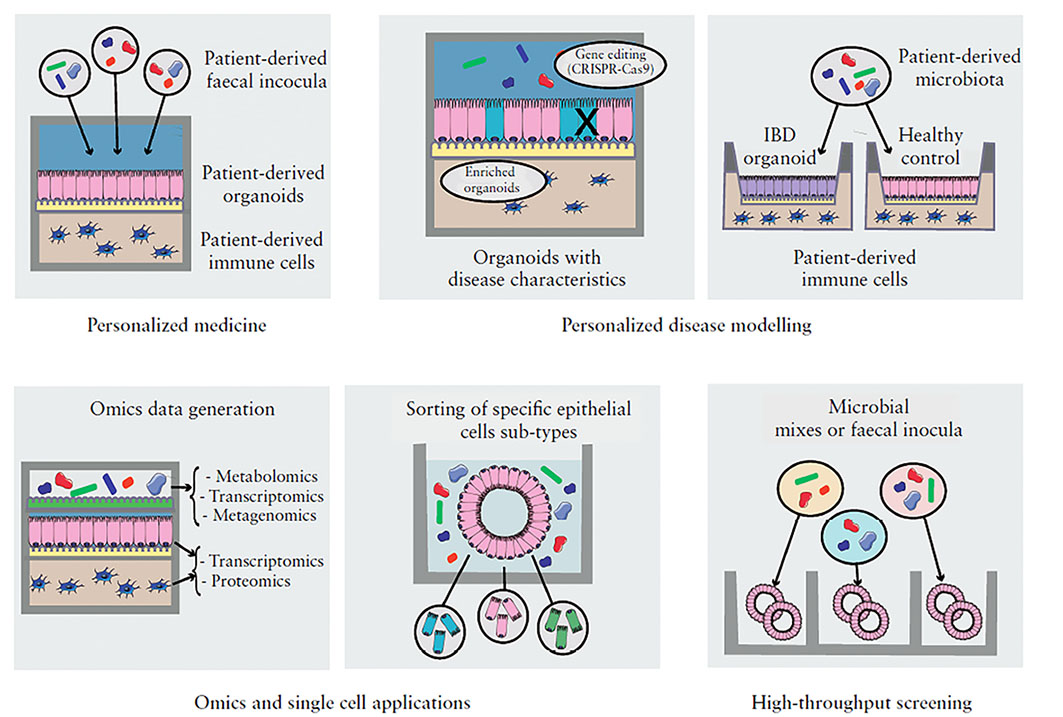
Figure 2: Organoid-based models applications. Source: Poletti et al, Journal of Crohn's and Colitis, 2020
For full-sized version, please click on the image.
Host cells and microbial gut communities present key inner and mutual interactions in health that are perturbed in digestive diseases, and we chose to investigate these disturbances from two complementary angles:
1) The human gut commensal bifidobacteria have been highlighted as protective agents against a number of disease conditions. Bifidobacteria can interact with host intestinal cells and influence their specific functions. Human intestinal organoids exposed to Bifidobacterium are being used to characterise the role of this commensal in the regulation of host functions in different epithelial cell populations. High-content bioimaging screening for autophagy proteins for example as well as omics data are being generated from exposed human colonoids to understand how autophagy is modulated. Integration of omics data with available published interaction data sources will help generate bifidobacteria-host molecular networks on a cell-type specific level, the analysis of which will identify key pathways (including autophagy) and functions that can be modulated by the bacteria.
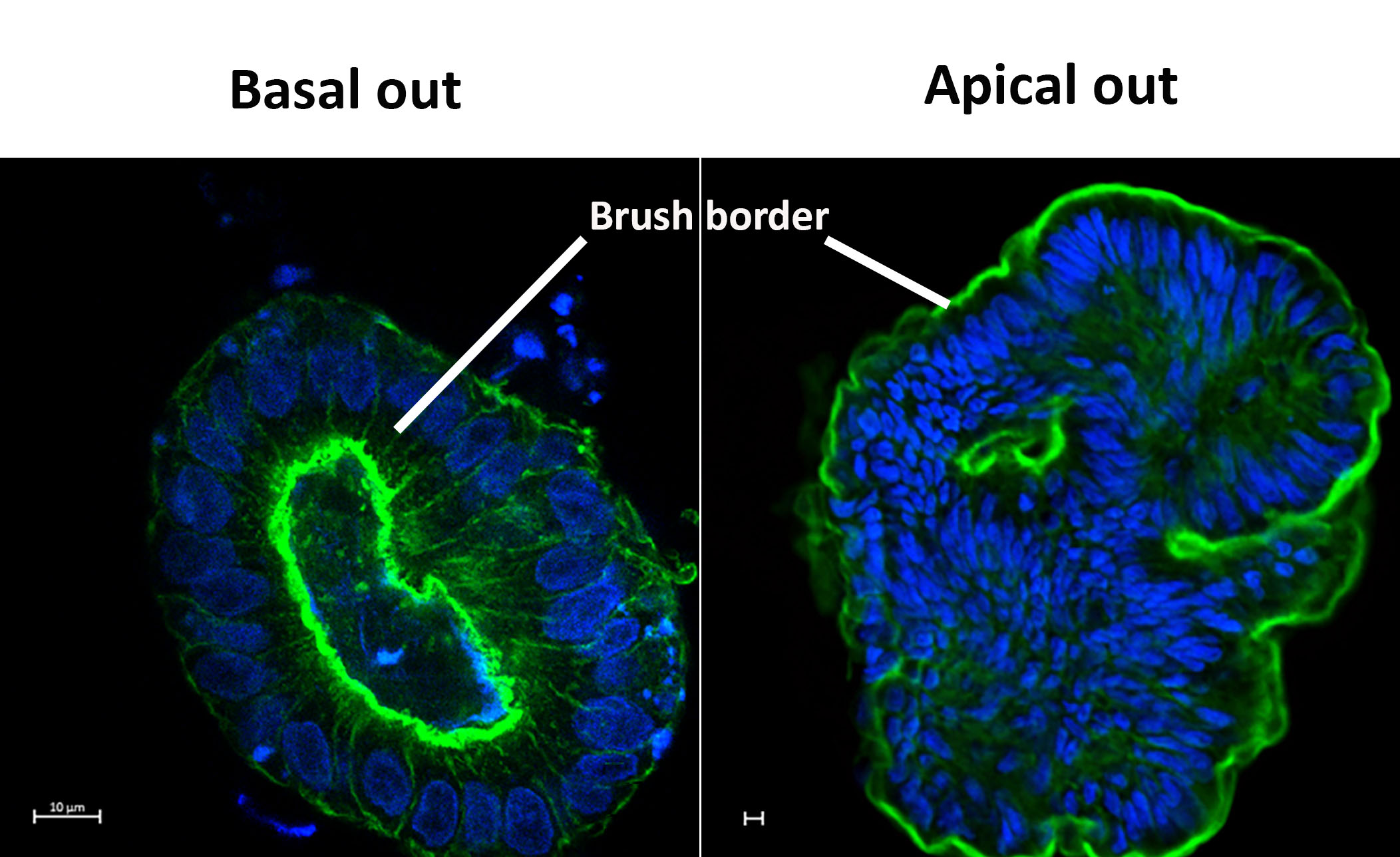
Figure 3: Basal out and apical out 3D human colonoids. Actin (Green) and Nuclei (Blue). Credit: I. Hautefort & M. Poletti
For full-sized version, please click on the image.
2) In pathologies such as IBD, many of the components of the gut environment besides the microbiota present severe alterations:
2.1) Among those, the bile acid metabolism is impaired and this has been shown to play a detrimental role in IBD pathogenesis. Unfortunately, the molecular and regulatory mechanisms by which bile acids maintain a healthy gut epithelial surface are not fully characterised, impeding the development of targeted therapeutics to restore normal bile acid metabolism when dysregulated like in IBD.
2.2) In parallel, a key cellular compartment of the gut, the enteric nervous system (ENS), displays tremendous structural and functional alterations in both Crohn’s disease and Ulcerative colitis, the two forms of IBD. Damage to the ENS cellular components (e.g. enteric neurons and glial cells) has dramatic repercussions on gut motility, blood flow, and epithelial barrier functions and innate defense (e.g. tight junctions, mucus productions, maintenance of the stem cell niche). Using human colonoid models, we are investigating the impact of selected primary and secondary bile acids, or key ENS secreted neuromediators on the regulation of cell type-specific functions and epithelial layer integrity. Furthermore, we are investigating how inflammation impairs the homeostatic effects that bile acids and ENS signalling molecules have on the gut epithelium. For this, we are using inflamed in vitro colonoids to identify host epithelial cell pathways targeted by bile acids and neuromediators which are altered during the onset of inflammatory diseases.
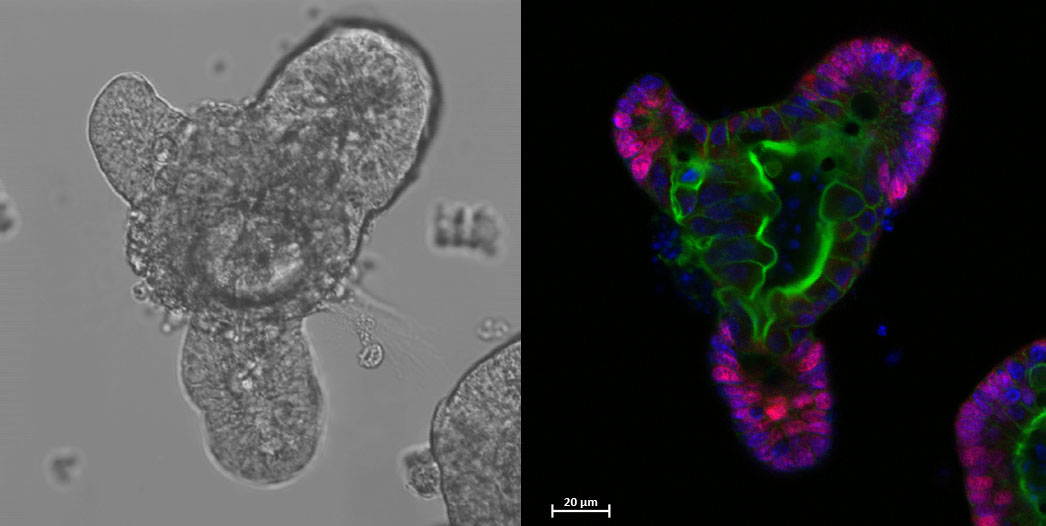
Figure 4: 3D mouse ileal organoid depicting proliferative regions (Pink), actin cytoskeleton (Green) and nuclei (Blue). Credit: I. Hautefort
For full-sized version, please click on the image.
Our organoid work is supported by the NIHR Imperial BRC Organoid Facility.