Project – Systems-level Projects on COVID19
The SARS-CoV-2 pandemic has mobilised scientists around the globe to research all aspects of the coronavirus virus and the disease it causes (COVID-19). Although the pathogen primarily targets the respiratory system, its involvement in other major organs has now been documented thoroughly.
To study COVID-19, we applied our expertise in multi-omics network modelling and re-utilised our resources. In particular, we have investigated how various infected cell types, especially in the gut, react to the pathogen, and how the immune response given to SARS-CoV-2 differs from other viruses Olbei et al, Frontiers in Immunology, 2021. We have also developed multiple new computational tools, such as ViralLink and CytokineLink, aimed at helping the scientific community study the disease (Olbei et al, Cells, 2021, Treveil et al, PLOS Computational Biology, 2021).
We combined all these network tools with single-cell data from SARS-CoV-2 infected ileal and colonic organoids as well as from intestinal immune cells from healthy patients. With this analysis, we reconstructed a map on how infected and bystander epithelial cells could influence gut resident immune cells. The methodology (that can be applied to other tissues and datasets) pointed out key intestinal epithelial ligands that could contribute to a systemic cytokine storm (Poletti et al, NPJ Systems Biology and Applications, 2022).
During these trying times we have made several strategic collaborations with other specialists from relevant fields, further emphasising the collaborative efforts needed to tackle global challenges such as the COVID-19 pandemic. We are actively contributing to the COVID Disease Map effort, a consortia of hundreds of researchers developing systems-level maps of SARS-CoV-2 (Ostaszewski et al, Molecular Systems Biology, 2021).
ViralLink: An integrated workflow to investigate the effect of SARS-CoV-2 on intracellular signalling and regulatory pathways
We have developed an integrated network analysis pipeline, ViralLink, intended to study viral infections in a cell-type specific manner, using transcriptomics data to provide mechanistic insights into the intracellular molecular interactions of SARS-CoV-2 infected cells. ViralLink is an easily accessible, reproducible and scalable systems biology workflow to reconstruct and analyse molecular interaction networks representing the effect of the viruses on intracellular signalling (Github). We believe it is the first available integrative workflow for analysing the downstream effects of viral proteins using viral host interactions and host response data, that can be applied to study the effects of any other viral pathogens as well. (Treveil et al, PLOS Computational Biology, 2021).
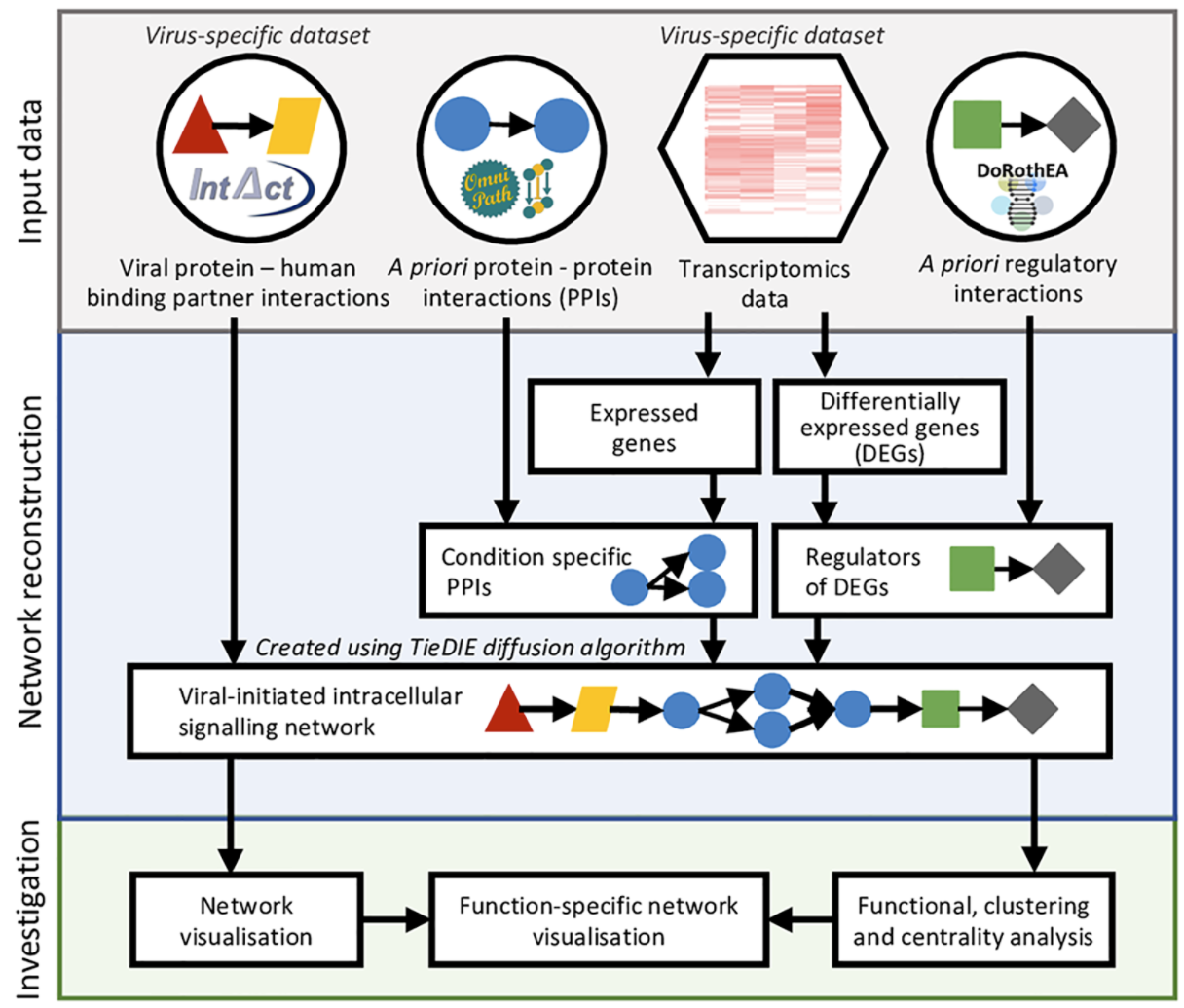
For full-sized version, please click on the image.
SARS-CoV-2 Causes a Different Cytokine Response Compared to Other Cytokine Storm-Causing Respiratory Viruses in Severely Ill Patients
Through a systematic literature curation effort, we identified shared and unique cytokine responses absent following viral infections given to cytokine release syndrome (CRS) or cytokine storm causing viruses, including SARS-CoV-2. Through the analysis of thousands of published research articles on patients infected with CRS causing viruses (H5N1, H7N9, SARS-CoV, MERS-CoV), we were able to identify a characteristic dysregulation of the type-I interferon (IFN) response, and highlight that the levels of specific cytokines did not increase in COVID-19 patients when compared with the responses of patients suffering from other viruses. Olbei et al, Frontiers in Immunology, 2021.
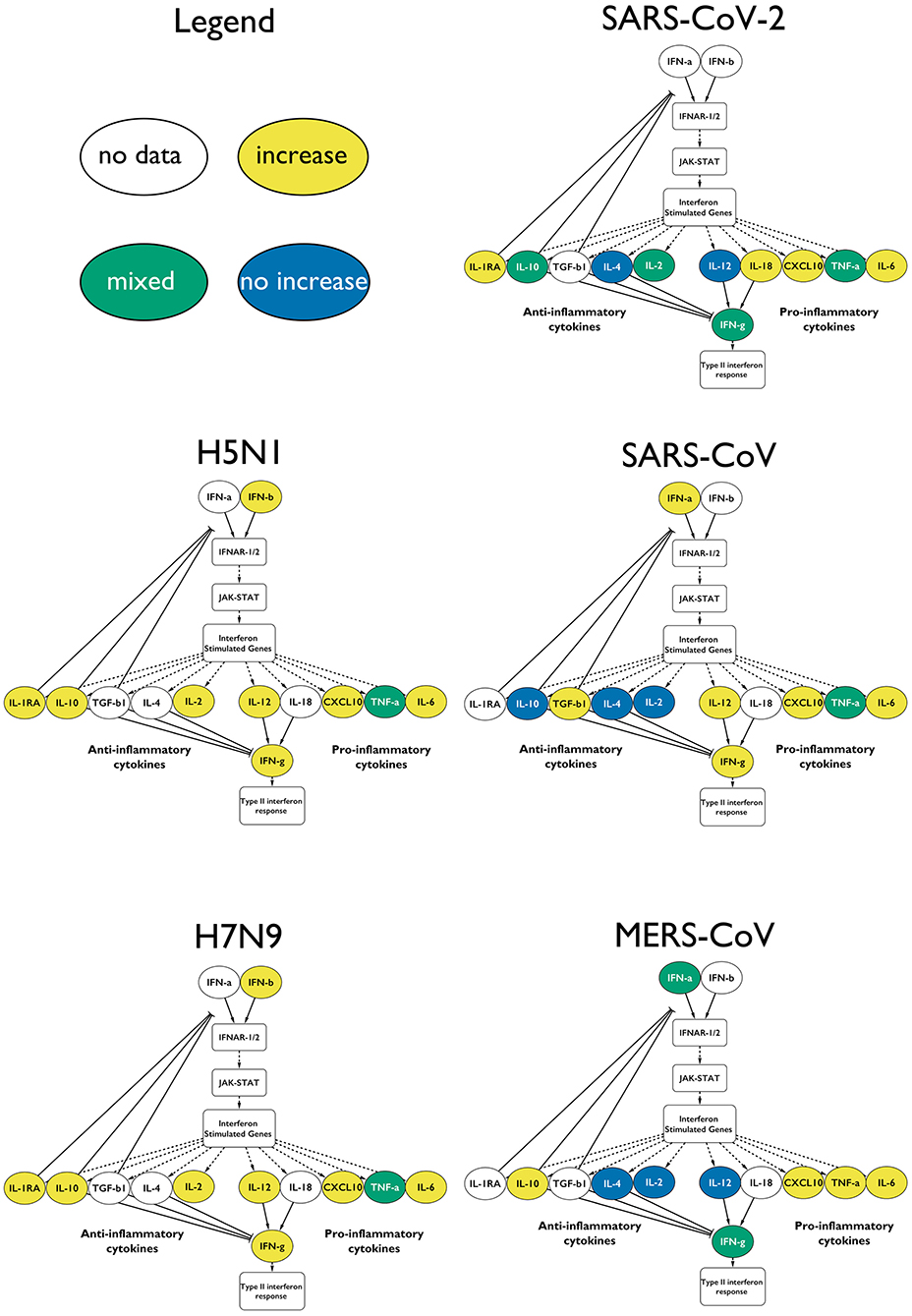
For full-sized version, please click on the image.
CytokineLink: A Cytokine Communication Map to Analyse Immune Responses
To understand how intercellular communication is mediated by cytokines during the development of immune responses, we developed a novel network resource, CytokineLink (Github). Through the integration of annotation data from multiple systems immunology databases, and introduction of a novel meta-interaction depicting how the binding of cytokines to their respective receptors might contribute to the release of other cytokines, we have provided a freely available network resource for the scientific community to study inflammatory and infectious diseases, such as Inflammatory Bowel Disease (IBD) or COVID-19, respectively. Olbei et al, Cells, 2021.

For full-sized version, please click on the image.
Mapping the epithelial-immune cell interactome upon infection in the gut
To gain insight into the role of the epithelium in the systemic and over-activated immune response to SARS-CoV-2 infection, we proposed an integrated computational biology framework, which models how infection alters intracellular signalling of epithelial cells, and how this change impacts epithelial cells and other local cell populations (https://github.com/korcsmarosgroup/gut-COVID).
Integrating transcriptomics data from SARS-CoV-2 infected human ileal and colonic organoids with intra-and intercellular network data, we were able to characterise the modified epithelial-immune interactome, pointing out specific interactions driving inflammation during disease response. Poletti et al, NPJ Systems Biology and Applications, 2022.
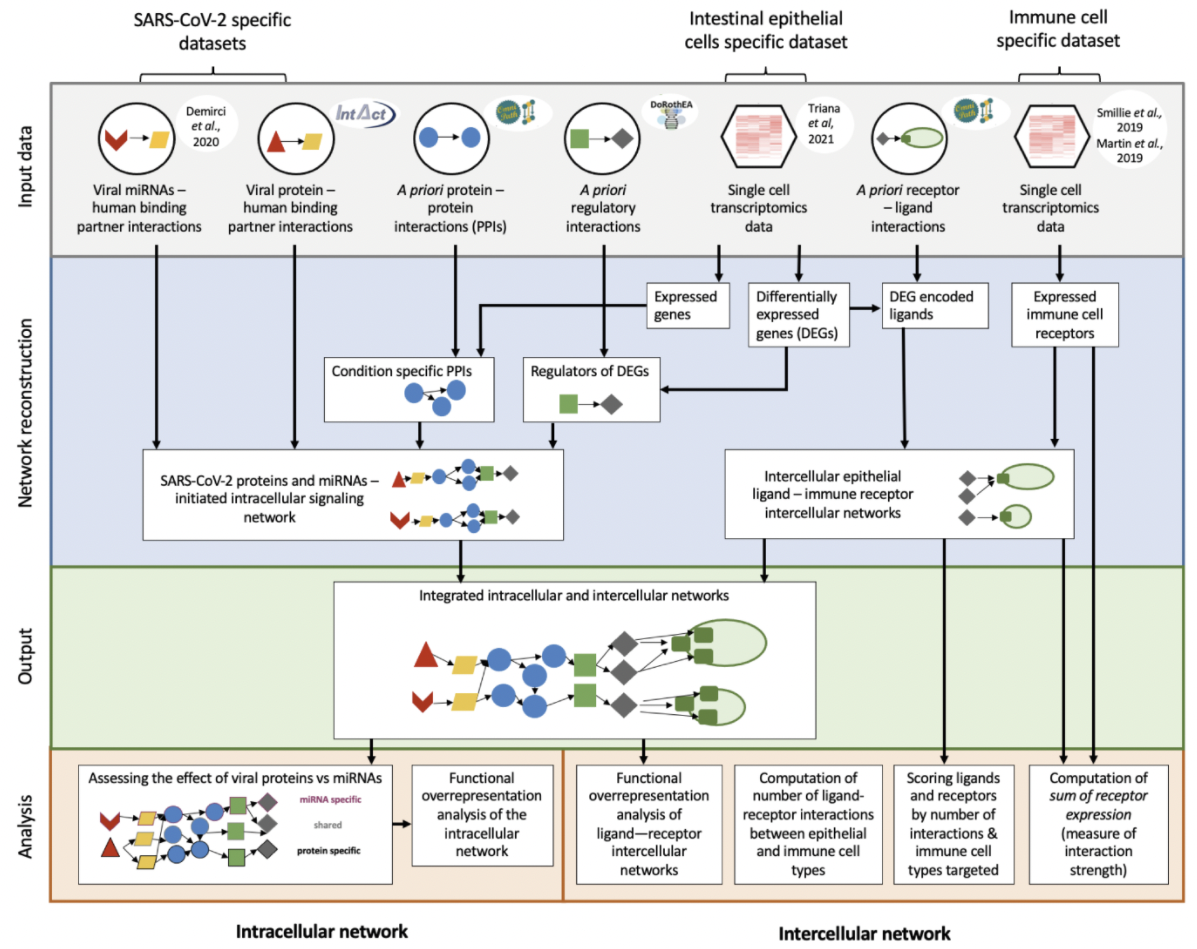
For full-sized version, please click on the image.
COVID-19 Disease Map, a computational knowledge repository of virus-host interaction mechanisms
Throughout the development of the previously listed studies, our group has actively contributed to the COVID-19 Disease Map (C19DMap) effort, a large-scale community effort to build an open-access interoperable and computable repository of COVID-19 molecular mechanisms. This massive undertaking combines tools, platforms and guidelines for graph-based analyses and disease modelling from hundreds of collaborators from 120 institutions all over the world.
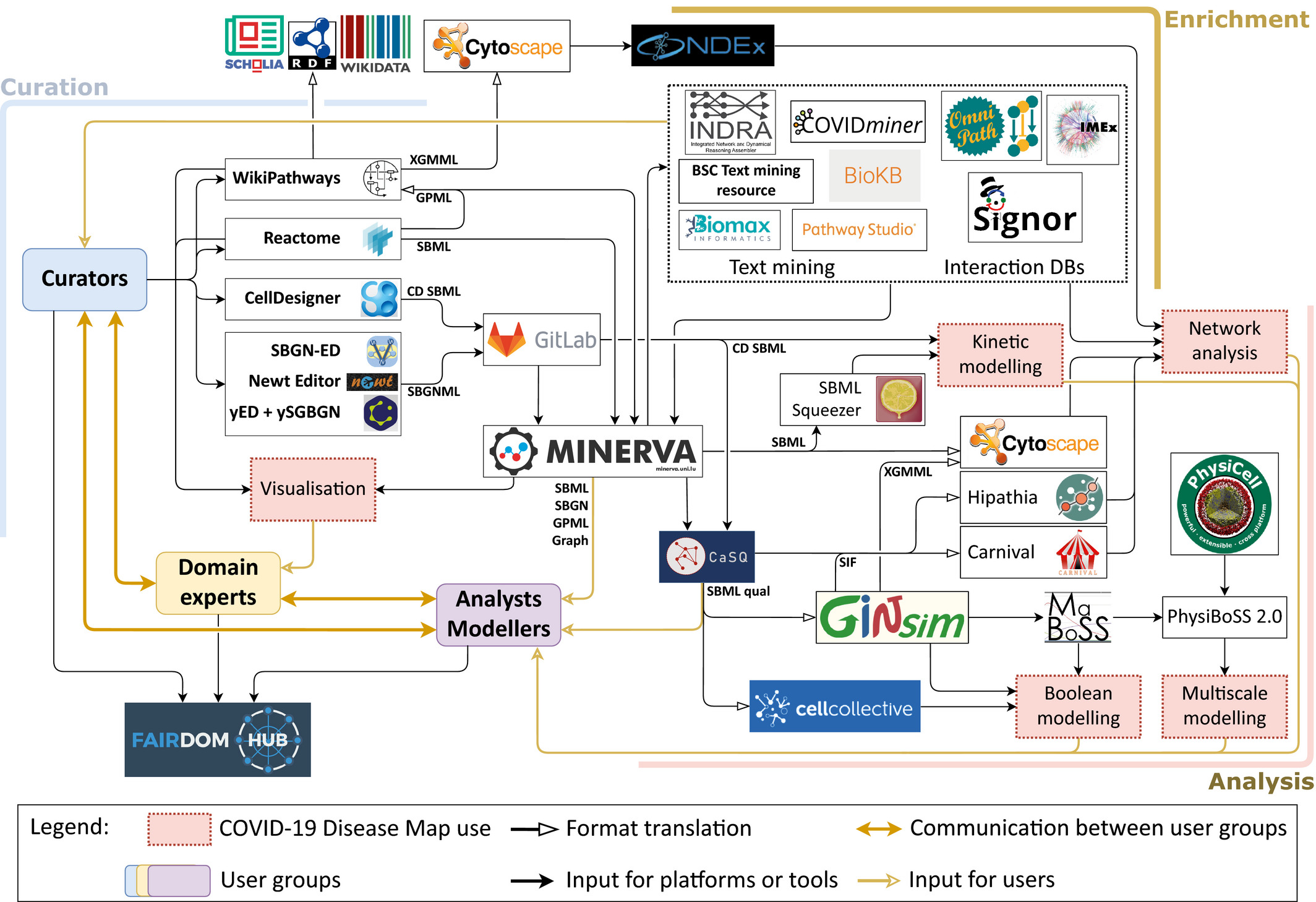
For full-sized version, please click on the image.